DNA Structure And Its Relation To Information Theory

The double helical structure of DNA is itched in everyone’s mind today. But most folks are not aware of the significance of this structure in functioning of DNA. In this post, I give you a simple explanation. DNA is the medium that biological beings use to store genetic information, and the features of DNA’s structure are what make this possible. I’ll try my best to keep the language simple enough for anyone to understand, regardless of their area of expertise.
Every advanced system, be it a book, a computer, or a biological system, relies on two main components- a method to store information, and a set of rules to interpret that information. In the English language, information is stored in the form of different combinations of 26 letters from A to Z. And a set of rules decides which combination of these letters has what meaning. This is how words and sentences are formed. When we learn English, we are basically learning these rules.
Computers use binary language. Instead of various combinations of 26 letters, they use various combinations of just two values, 0 and 1, to store and process information. And the set of rules used to interpret these combinations of 1s and 0s is called ASCII, or American Standard Code for Information Interchange.
Biological organisms, be they bacteria, plants, fish, birds or humans, use various combinations of 4 different molecules, namely adenine, thymine, guanine and cytosine, to store and process information. These four molecules are referred to by their first letters, as A, T, G and C respectively, and they are collectively called nitrogen bases. And the set of rules to interpret this information is called the genetic code. That’s something I will come to later in this article. This information in the form of various combinations of A, T, G and C is stored in DNA. And I will now explain how that works.
Structure of DNA
Before being incorporated into DNA, adenine, thymine, guanine and cytosine are converted into adenosine monophosphate (AMP), thymidine monophosphate (TMP), guanosine monophosphate (GMP) and cytidine monophosphate (CMP) molecules, respectively. These four molecules are collectively called nucleotides, and they are ever-present in a cell, suspended in its protoplasm, which is the liquid inside the cell.
A nucleotide is made by combining three molecules- deoxyribose sugar, phosphoric acid and nitrogen base, which can be A, T, G or C. Deoxyribose sugar gets its name from the fact that its second carbon atom lacks an OH group. And this is where the DNA gets the ‘Deoxyribo’ part of its name. And phosphoric acid is what makes DNA itself an acid. The ‘Nucleic’ part of the DNA’s name simply comes from the fact that DNA resides in the cell’s nucleus. And the fact that DNA is an acid makes the nucleoplasm (the DNA containing liquid inside nucleus) slightly acidic.
The structures of nitrogen bases and nucleotides are shown in the figure below. On the right are the four nitrogen bases. Uracil is a fifth nitrogen base, which is found not in DNA but in a related molecule called RNA. On the left is the structure of nucleosides made by combining a ribose (in case of RNA) or deoxyribose (in case of DNA) sugar with either one, two or three molecules of phosphoric acid. When one of the nitrogen bases is linked to the first carbon atom of the deoxyribose sugar, the nucleoside turns into a nucleotide, the building block of DNA.
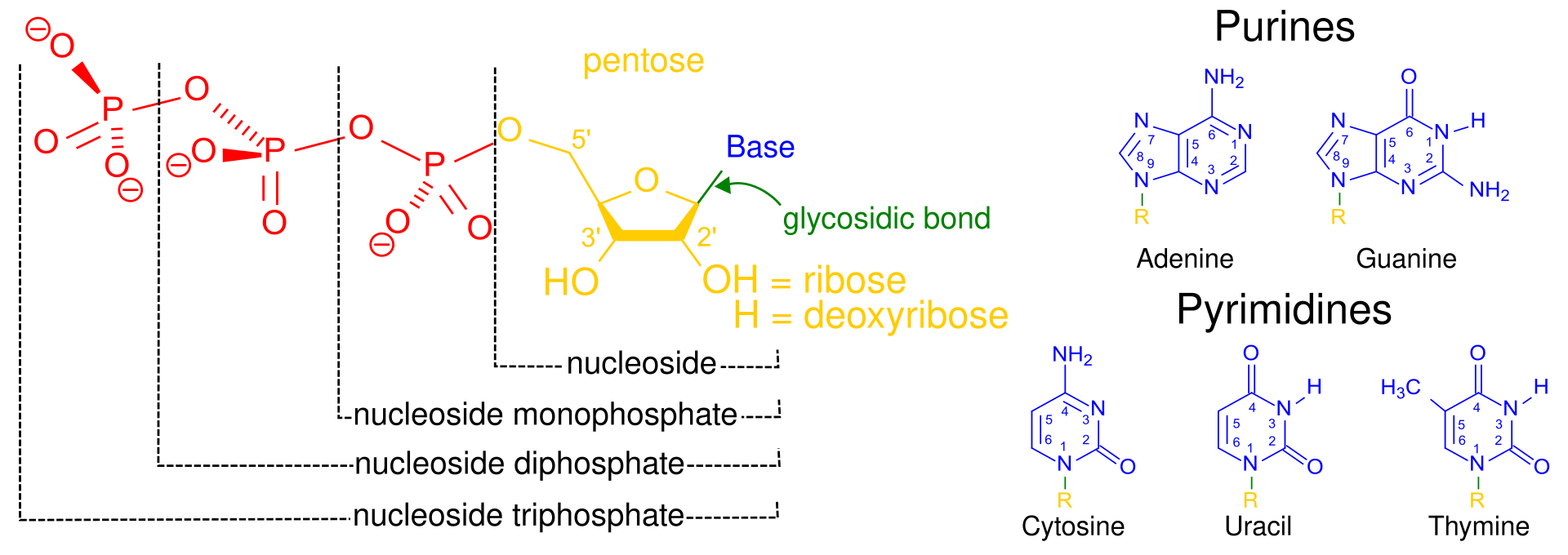
Now, DNA consists of two chains, or strands, twined around each other to form the double helix that the world is now familiar with. The figure below shows the structure of these strands. Each strand is made by joining various nucleotides through covalent bonds in linear fashion. The covalant bond forms between the OH group of carbon number 5 of the deoxyribose sugar part of one nucleotide, and one of the oxygen atoms of the phosphate group of the next nucleotide.
In this way, the chain contains a long sequence of the four types of nucleotides covalently bonded. And information is stored in the order or combination of the nucleotides in this sequence. It is this random sequence of A, T, G and C, like AATTGCTACC, that encodes every genetic trait of yours, like the shape of your nose, the colour of your eyes, how susceptible you are to some disease, how allergic you are to some allergen, how tall you are, how dark or fair you are, and possibly even some aspects of your personality.
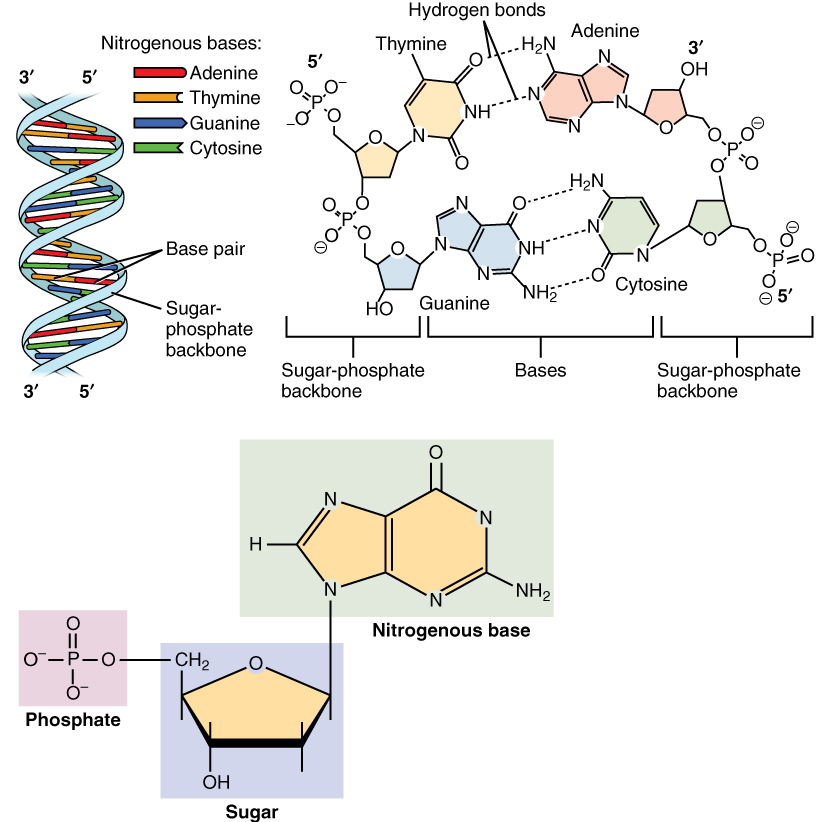
But DNA, as is common knowledge, has not one but two intertwined strands. Only one of these strands stores genetic information. This strand is called sense strand. And the other strand, while also made of nucleotides, doesn’t store any genetic information. This strand is aptly named nonsense strand. The nonsense strand has a sequence of nitrogen bases that is determined by the sequence of sense strand by the following rules. Where there is an A in sense strand, there is T opposite to it in the nonsense strand and vice versa. And where there is G in sense strand, there is a C opposite to it in the nonsense strand and vice versa.
This happens because as the figure above shows, adenine and thymine form two hydrogen bonds with each other, and guanine and cytosine form three hydrogen bonds with each other. But hydrogen bonding between A and G, or A and C, or G and T or T and C, is not energetically favorable. It is this hydrogen bonding between nitrogen bases of the two DNA strands, that keeps them together to form the double helical DNA molecule we all know. Two nitrogen bases in opposite strands hydrogen bonded to each other are collectively called a base pair.
Within an individual strand, there are also stacking bonds or pi bonds between the aromatic rings of adjacent nitrogen bases. There is another property of base pairs that determines the structure of DNA. A and G are roughly the same size, collectively called purines, and T and C are roughly the same size, collectively called pyrimidines. Hence a base pair, be it GC or AT, has roughly the same length. This gives the DNA molecule a uniform diameter of 2 nanometers throughout its length, regardless of its nucleotide sequence.
Packaging of DNA
Consider this for a moment. The nucleus of an average human cell measures just 10 micrometres in diameter, and yet contains DNA with a total length of around 2 metres! I am sure you are familiar with earphone wires getting tangled and had a hard time unravelling them. That should give you an idea of the enormous challenge a cell’s nucleus faces in accommodating the 2 metre long DNA in its tiny space and ensuring that it doesn’t get tangled.
To solve this problem, cell has come up with a device called nucleosome. DNA doesn’t just exist as a free thread. Nucleosomes are cylindrical structures made of histone protein molecules, around which the DNA is wrapped. This is shown in figure below. These nucleosomes basically serve the same function that a spool serves. Like a thread is wound around a spool to keep it neatly packaged, DNA is wound around nucleosomes, nature’s own nanospools.
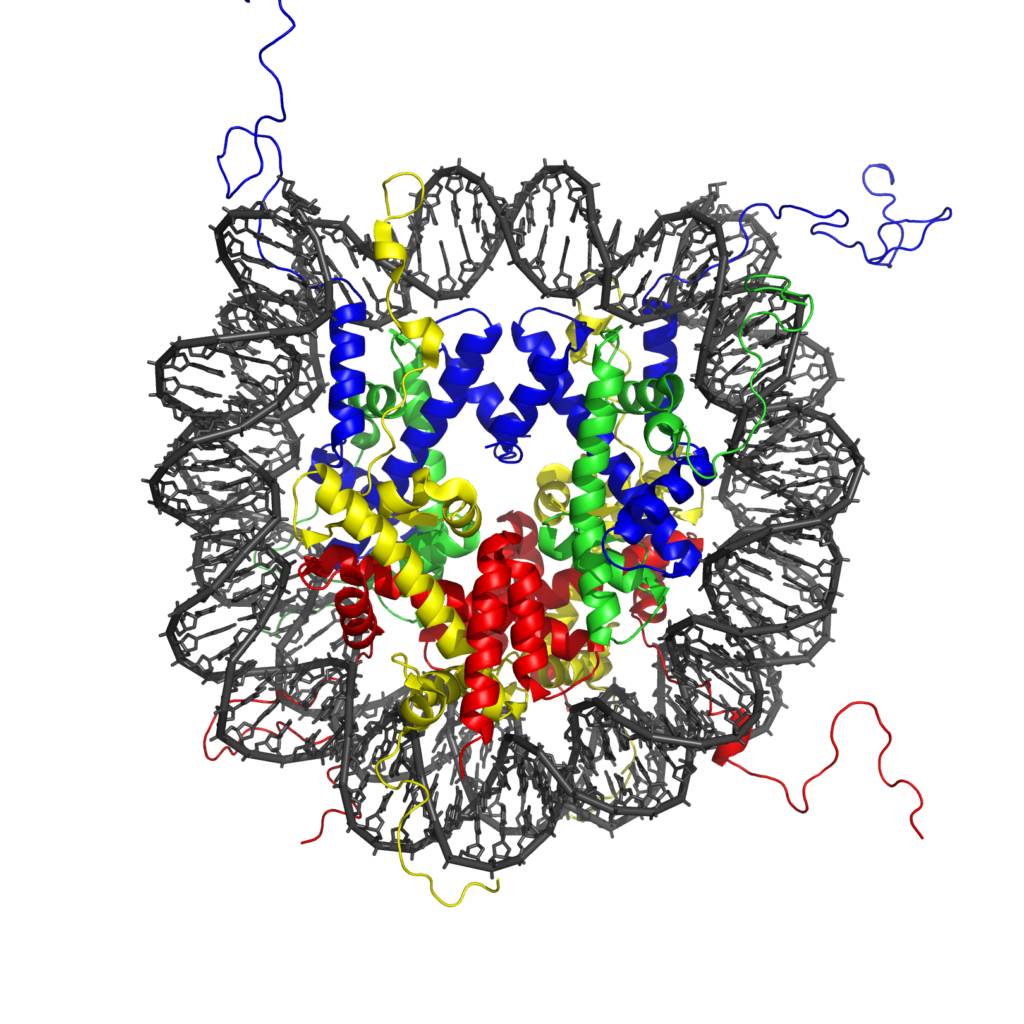
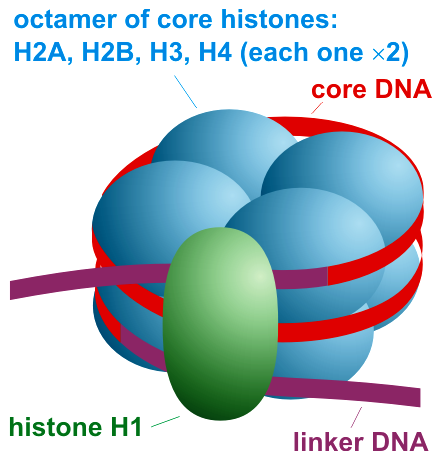
The reason why DNA wraps itself around a nucleosome is that while the phosphate groups of DNA have negatively charged oxygen atoms, the histone proteins that make up the nucleosomes have many positively charged amino acid residues on their surface. These opposite charges create attraction between DNA and nucleosome. One nucleosome has approximately 1.6 turns of DNA wrapped around it. Normally, while DNA is wrapped around a series of these nucleosomes, it is still loosely spread around the nucleus to enable the cell’s nano-machinery to read it’s information.
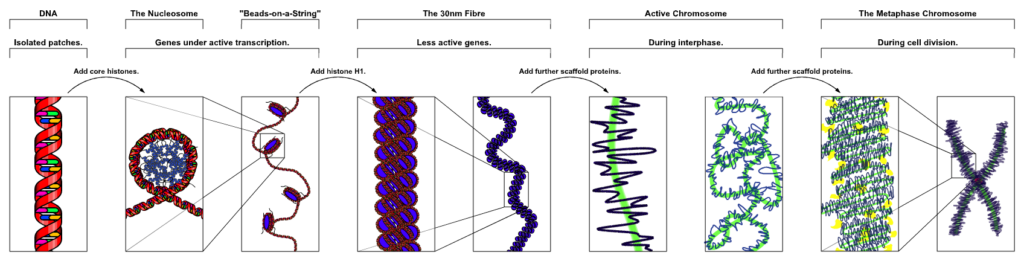
But during cell division, the nucleosomes are packed successively into a series of higher order helical structures as shown in figure below, ultimately leading to the formation of chromosome. Think of this as further organising of multiple spools into a spool rack. This is actually an example of fractal structure. Fractal structures involve self similarity at different scales. In this case, you see helices at different levels or scales.
At the lowest scale, DNA itself is a double helix. On the next level, DNA wraps itself around a nucleosome in 1.6 turns, creating another helix. Then in the next level, the nucleosomes connected by the DNA wrapped around them, themselves fold into a compact helix, which then further folds into an even higher order helix, and so on. This is what allows packaging of a 2 meter long DNA molecule into chromosomes whose sizes can be measured in nanometers. These neat and compact chromosomes can then be easily transported to the opposite ends of the dividing cell by the cell’s molecular machinery.
DNA and Information Theory
At this point, its useful to look at what exactly information is, and what determines how useful it is. Let’s first understand another concept- entropy. Entropy is nothing but the amount of disorder in a system. A set of randomly scattered crossword pieces has more disorder, and hence more entropy, than a set of crossword pieces arranged to form a word, which in turn has more entropy than just letter A crossword pieces neatly arranged in a row.
But out of these three sets, the one most useful to us is the one arranged to make a word. But notice that this is not the set that contains most information. The set that contains the most information is the one in which the letters are randomly scattered, the One with the most entropy. This is because information is nothing but entropy. The higher the entropy in a system, the higher is the amount of information in it. The following example will illustrate this.
Suppose that the group of pieces form the word queen. This word can be conveyed as qu—n, and anyone will guess that it means queen. This is because the chance of qu being followed by ee is very high in English language. Hence, to store the word queen, you just need to store the sequence qun in order for it to make sense. This is, in fact, one of the things that enables compression of computer files, reducing file size. However, if the pieces are just randomly arranged in the sequence dgfjsg, you need to store the entire sequence, as you can’t predict what letter follows any of the letters in it.
In other words, you can’t compress it. This shows that queen contains less information than dgfjsg, even though they are both made of English letters. And yet, its the lesser information of queen, and not the more information of dgfisg, that is useful to us. This shows that the very act of using a medium for useful communication reduces its entropy, or information carrying capacity.
This is because to use the medium for communication, you need to device a set of rules. These set of rules can be the grammar of a spoken language, the ASCII code of binary language, or the genetic code by which DNA stores genetic information. But a set of rules, by its very nature, puts constraints on the freedom of the medium. No longer can the letters of a language be arbitrarily arranged. And these constraints reduce entropy and information in the medium. It is this reduced information, however, that can be used as a language for communication.
This reduction in information due to an attempt to use the medium for communication enables software files to be compressed, and enables even incompletely written English words to be understood. But is it also observed in the information coded in DNA? Yes, it is. At several levels in fact. On the first level it is observed when we look at a chromosome as a whole.
There is a stain called Giemsa stain, made by mixing methylene blue, eosin and Azure B. When DNA is exposed to Giemsa, the stain molecules specifically bind to AT base pairs in DNA and not to GC base pairs. So, the more the number of AT base pairs in a stretch of DNA, the more heavily that stretch of DNA will be stained.
Now, DNA can carry maximum information when its entropy is maximum, as information is nothing but entropy. And entropy of DNA will be maximum when the AT and GC pairs are evenly distributed through out the DNA molecule. And if this is the case, then the DNA molecule will be uniformly stained by Giemsa through its length. In such a case, the chromosome stained with Giemsa would look uniformly grey all through its length. But in reality, chromosomes stained with Giemsa look like in the image below.

Instead of being uniformly grey, they have certain regions that are black, or very heavily stained with Giemsa, and other regions that are almost white, or hardly stained at all. This shows that instead of AT and GC base pairs being evenly distributed through out the length of the chromosome DNA, some regions in DNA have many more AT pairs than GC pairs and other regions have many more GC pairs than AT pairs.
Typically, the AT rich regions don’t contain any genes and are called heterochromatin, while the GC rich regions contain genes and are called euchromatin. This means that the amount of information these genes can carry is less than the amount of information DNA can theoretically carry with maximum entropy, as less AT base pairs would be available to code information for the genes in GC rich euchromatin regions.
Second level in which redundancy and reduced information is seen is when we compare the nucleotide sequences of different genes. A particular sequence in a gene can be used to predict the sequence right ahead of it, because the probability of the first sequence being followed by the second sequence is higher that what would be if the base pairs were randomly distributed. Notice the similarity in this case, to the high probability of q being followed by ueen in English. In fact, this ability to predict the sequence of nucleotides ahead of the short sequence you know is very useful in analyzing and comparing genes using bioinformatics software.
A third level at which redundancy is observed is in the genetic code itself. The figure below shows the genetic code. In genetic code, there are a number of codons, each codon composed of three nitrogen bases. And each of these codons codes for a unique amino acid. Without going into too much detail, a brief explanation is that the sequence of nucleotides in DNA determines the sequence of codons, and the sequence of codons in turn, determines the sequence of amino acids in the protein that the gene is producing.
Proteins are nothing but many amino acids linked to form a chain. There are 20 different amino acids that can be incorporated into proteins, and every protein has a unique amino acid sequence, which makes it chemically and structurally different from other proteins. And this amino acid sequence of the protein is determined by the nucleotide sequence of the gene producing it.
Now, as the figure below shows, there are 64 different codons made from the A, T, G and C nucleotides, each containing three nucleotides. So theoretically, The genetic code should be able to encode 64 different amino acids, with each codon encoding for a unique amino acid. But instead, as the figure shows, multiple codons encode for the same amino acid, causing only 20 different amino acids to be encoded by the genetic code.
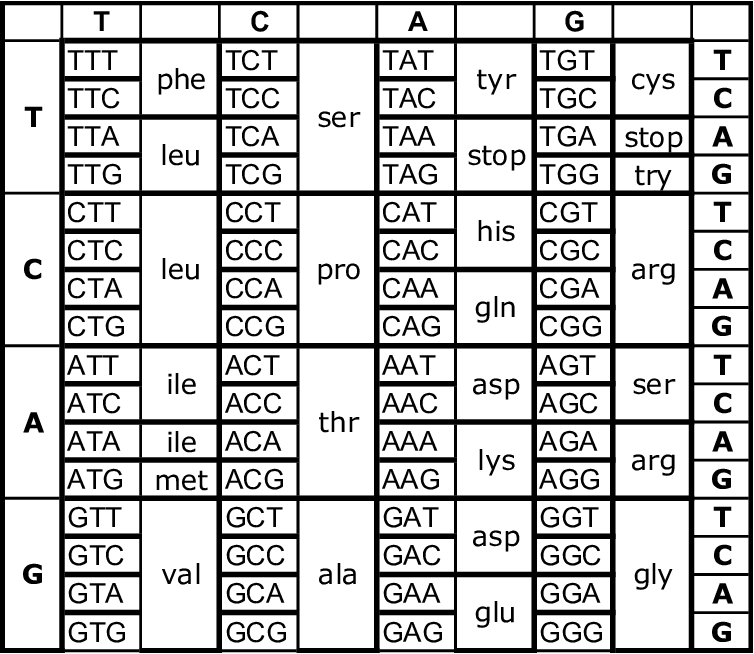
This is yet another example of redundancy. And this again enables you to predict amino acid incorporation. For example, all four codons starting with CT sequence code for the same amino acid, namely Leucine (leu), regardless of which nucleotide lies in the codon’s third position. So even if you don’t know the identity of the nucleotide in the third position, you can guess that the codon codes for leucine if its first two nucleotides are CT.
Why this redundancy has evolved in DNA on so many levels is a tricky question to answer. But you already know the underlying reason. Any attempt to use a medium to store information imposes a set of rules on it, which invariably reduces its entropy, and therefore, information carrying capacity.
Speaking specifically of the genetic code, incorporating even 20 different amino acids in proteins based on a predetermined plan requires the cell to have a molecular machinery of bewildering complexity. Incorporating 64 different amino acids would probably have taxed the cell’s resources to the breaking point.
But regardless of the extent to which the information carrying capacity of DNA is reduced due to the sets of rules imposed on it for storing genetic information, DNA is still capable of carrying all the information that decides pretty much every trait of yours, from your physical appearance, susceptibility or immunity to diseases, and even personality to some extent. It makes us who we are.